
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Help for Codon Optimization
Table of Contentsback to top 1: Submitting a JobThis website generates and visualizes a codon optimized nucleotide sequence based on the following inputs:
In the rest of this document, each given user-submitted sequence (and its input parameters) is referred to as a "job". The overall process of running a job can be summarized as follows:
Further details on each step or input above, may be found in the sections below. Other pages on this website also include a "help" link at the top right corner, which directs you to its respective section within this guide. If you experience trouble using this site, or have other comments and suggestions, feel free to contact us. Important: All the setup pages have "Save and Continue" near the bottom left corner. You must click on this for your inputs to be saved. Navigating to the other setup pages, by clicking on their link tab along the top row, will bring you there without saving any changes that you may have made. On the other hand, if you do NOT want to save changes you may have made (e.g. if you accidentally erased some previously saved input), then navigating to another page (by clicking on their link tab along the top row without clicking "Save and Continue"), will discard your changes. back to top 1.1: Input SequenceHere, you enter the desired sequence which you wish to optimize. You first select whether you wish to input protein or DNA/RNA sequence. Each has a seperate set of format requirements, which is noted just below the sequence type name. For testing purposes, there are links to the right of the sequence type name. When you click on them, it will open a sample sequence of that type, in a new window. If you wish, you may copy and paste this sample sequence into the input textbox. For testing purposes, there are buttons to the right of the sequence type name. When you click on them, it will load a sample dataset into the input sequence textbox, and then automatically Save and Continue. These buttons require Javascript to work, and may fail to function if Javascript is not fully enabled. Important: These buttons will overwrite your current input sequence (if any). If your input is DNA/RNA, you also need to select which set of translation rules applies to your nucleotide input. Important: This translation rules you select here, applies to the source organism of the nucleotide sequence, where the DNA/RNA was taken from. Later on, under 2: Optimization Settings, you may select another translation rule which applies to the expression host instead. E.g. lets say you are trying to express a Blepharisma nucleotide sequence in Salmonella bongori (which uses Standard translation rules). Then here, you should select "Blepharisma Nuclear Code". Optionally, you may also enter a title for the current sequence, which will be included in the output results. This might be useful, if you are submitting several jobs at once, and you wish to keep track of which result belongs to which job. When you are done, click "Save and Continue". If you are initially submitting a new sequence, the link tabs for the other steps (2-5) are greyed out and inactive. These other setup pages only become available, after you first save a sequence. After the initial step of saving a sequence, but before you Submit the job, you may still return to this "1: Input Sequence" page to edit your sequence and/or title. If you are just submitting a job for testing purposes, note that this is the ONLY page that requires user input. The rest of the pages either provide options which come with some reasonable default selection (e.g. target species is initially set to Escherichia coli by default), or have inputs which are entirely optional (e.g. exclusion nucleotides). Hence, beyond this page, for the rest of the job submission process, you can just click "Save and Continue" without changing anything, all the way until the job has been submitted. back to top 1.2: Optimization SettingsThere are several options to set here. Firstly, you need to select your optimization criteria. There are 4 parameters, and each parameter has 3 settings: "Ignore", "Maximize" and "Minimize". At least 1 parameter should be selected (i.e. set to Maximize or Minimize) in order for the Algorithm to have some optimization target to work with. Individual Codon Usage: abbreviated as ICU. This refers to how closely the output nucleotide sequence matches the codon usage pattern of the Expression Host. When set to "Maximize", the output sequence will try to match the host as closely as possible (subject to other optimization criteria, and motif settings). Conversely, when set to "Minimize", the output sequence codon usage pattern will try to deviate from the host as much as it can. Codon Context: abbreviated as CC. This is like ICU, but applied to pairs of codon (6 nucleotides in all), instead of each individual codon. Hence when set to maximize, it tries to match the codon pair usage pattern of the Expression Host. The recommended default criteria is to Maximize Codon Context only. Codon Adaptation Index: abbreviated as CAI. It measures the fitness for each codon, relative to the target expression host, but it is calculated differently from ICU. The score for each position is calculated by using the host frequecy of the codon at that position, divided by the host frequency of the most common synonymous codon for that amino acid. (E.g. Phenyalanine is encoded by both TTT and TTC. Lets say that the expression host uses TTT with a frequency of 0.8, and TTC with a frequency of 0.2. The score for each occurence of TTT is 0.8/0.8 = 1, and the score for each occurence of TTC is 0.2/0.8 = 0.25). Hence, CAI is maximized, when only the most common codons are used. Number of Hidden Stop Codons: In each protein-coding sequence, there is only ever 1 stop codon, at the end of the protein. Instead, this parameter refers to stop codon sequences that are found outside the normal reading frame. (E.g. Methionine-Threonine is encoded by ATGACN, and the underline highlights a hidden stop codon). Selecting "Maximize" on "Number of Hidden Stop Codons", will maximize the number of hidden stop codons in the output sequence. This may be useful according to the ambush hypothesis, by causing erroneous frame-shifted translations to terminate early.
In the second section, you can enable Codon Auto-Correlation as an optimization parameter, and specify a target value. For each amino acid, all the codons for that particular amino acid are compared against one another (ignoring the codons for other amino acids which may occur in between). A target value of 1 maximizes the probability that the each synonymous codon will be the same as its immediate neighbour (e.g. for Cysteine, it might be UGC=UGC=UGC=UGC=UGC). A target value of 0 maximizes the probability that the each synonymous codon will be different from its immediate neighbour (e.g. for Cysteine, it might be UGC#UGU#UGC#UGU#UGC). A target value of 0.5 does not necessarily mean that each synonymous codon has a 50% chance of being the same as its immediate neighbour. Rather, it means that the distribution of similiar synonymous codons, would be the same as expected by random chance. This chance in turn, depends on the total number of possible codons which code for that amino acid. For each given pair of synonymous codons, the chance that they are the same is equal to 1/(total number of possible codons for that amino acid). E.g. Lets say there are 5 synonymous codons, which can be grouped into 4 sets of codon neighbours. If there are a total of 2 possible codons for that amino acid, then each codon has a 1/2 chance of being the same as its previous neighbour. This computes to an average of 4*1/2 = 2 neighbouring synonymous codons which match (e.g. for Cysteine, it might be UGC=UGC#UGU=UGU#UGC). If there are a total of 4 possible codons for that amino acid, then each codon has a 1/4 chance of being the same as its previous neighbour. This computes to an average of 4*1/4 = 1 neighbouring synonymous codons which match (e.g.for Glycine, it might be GGA#GGU#GGC#GGG=GGG) In the results, this target value will be used to calculate a Fitness value, i.e. how far the sequence deviates from the target. To calculate the Fitness for the whole sequence, we average the following fitness score for each amino acid [ Absolute( Match_Count / ( Match_Count + ( Mismatch_Count / (Number_of_possible_codons - 1) ) ) - Target_Score ) ]. We ignore amino acids which only have one possible codon, and amino acids which only occur once. In the thrid section, you can enable 5' RNA folding instability as an optimization target. If you maximize instability, the RNA is less likely to fold on itself, allowing ribosome the easier access for initiation. Because calculating folding energy for the whole RNA strand is time-consuming, we limit calculations to the first 10 to 50 bases on the 5' end, where translation initiation occurs. CoDOn uses the Vienna RNA package to calculate RNA folding energy:
In the fourth section, you can enable GC content as an optimization target. There are 2 seperate parameters here: Total GC content, and GC3 content. Total GC content refers to the GC content of the entire sequence. This field might be useful if your desired nucleotide sequence should have a specific GC content value (e.g. if it needs a specific melting temperature). To enable GC content optimization, ensure that the checkbox is ticked, and enter your target as a percentage value (e.g. "38" instead of "0.38"). GC3 content refers to the GC content of only the 3rd base of all codons in the sequence. The 1st and 2nd bases are completely ignored. (E.g. the sequence ATGTGGTAA will have a GC3 content of 2/3 = 66.67%.) To enable GC3 content optimization, ensure that the checkbox is ticked, and enter your target as a percentage value (e.g. "38" instead of "0.38"). Some references for GC3 are listed below:
The next section specifies the individual codon (and/or codon pair) usage pattern values, which will serve as the optimization targets for the expression host. Firstly, there is a radio button, where you choose either to "Use Inbuilt Expression Host", or to "Input Custom Codon Usage Pattern Values". If you choose "Use Inbuilt Expression Host", you are using values for one of our inbuilt species. You may select the target species from the dropdown list. In the next page, you can then pick genes of the selected Expression Host to use. The algorithm will also automatically apply the translation rules appropriate to that species (i.e. which codons code for which amino acids). If you choose to "Input Custom Codon Usage Pattern Values", then you need to select the translation rules for your custom species from the dropdown list. Important: This translation rules you select here, applies to the expression host, where the protein will be produced. Whereas the translation rules you selected back in 1: Input Sequence applies to the nucleotide sequence's original host, where the DNA/RNA was taken from. E.g. lets say you are trying to express a Blepharisma sequence in Salmonella bongori (which uses Standard translation rules). Then here, you should select "Standard Code". Additionally, you also need to input the individual codon (and/or codon pair) usage pattern values for whichever optimization criteria is being used. (E.g. if you chose to maximize or minimize "Individual Codon Usage" or "Codon Adaptation Index" at the top, you need to provide codon usage pattern values for your custom species in the "Individual Codon Usage Pattern" textbox.) There are 2 ways you can input the codon usage pattern values, as follows: The first way is to upload the coding sequences of your target host as FASTA format, and the website will convert these sequences into the appropriate codon usage pattern values. To do this, click on "Convert Fasta Into Codon Usage Pattern Values", and you will be led to another page, where you can upload your FASTA formatted file. If the upload is successful, your converted codon usage pattern values will be seen in the "Individual Codon Usage Pattern" and "Codon Context Usage Pattern" textboxes. The second way is to calculate the codon usage pattern values on your own, format these values into text, and paste the text into the provided textboxes. The text format for codon usage pattern values take the general form of a list of nucleotide sequences on seperate lines. Within the same line, each nucleotide sequence is followed by a space or tab, and then an integer stating the frequency of that sequence. Please only use integers, and not fractions or decimals. For Individual Codon Usage, the nucleotide sequence will be exactly 3 bases long (Sample Format). For Codon Context, the nucleotide sequence will be 6 bases long (Sample Format). When you are done, click "Save and Continue". back to top 1.3: Select GenesYou only come to this page, if you opted for "Use Inbuilt Expression Host" in the previous page (Optimization Settings). If you had instead opted to "Input Custom Codon Frequency Values", this page becomes unnecessary, and will be skipped. You will instead be sent directly to the next page (Motif Settings). Here you may select which genes of the Expression Host are included, when calculating the codon (and/or codon pair) frequency values, which will serve as the optimization target. By default (i.e. if you make no changes), we will use all the genes for the Expression Host. Genes here are always referred to by their Locus Tag ID. There are two ways by which you can select genes. At the top-most segment of the page is the "Input Quick Gene Selection List" textbox. If you have a list of genes you want to use (e.g. from sorted Gene Expression Data), simply copy and paste it into the textbox, and then click "Select Genes From Text Input", next to the textbox on the lower right. The website will then select all the genes which appear in the textbox, while de-selecting everything else. In the event that a gene ID is not recognized, it will be noted under the "Unrecognized Gene ID List" further below. Important: Unlike the other pages, "Input Quick Gene Selection List" is not saved by "Save and Continue". Instead, it is saved by "Select Genes From Text Input". The other way is to move genes between the "Selected Genes" and the "Genes Available for Selection" list. Note that you can select genes independently of the "Input Quick Gene Selection List" textbox. Hence you can ignore the "Input Quick Gene Selection List" textbox and just select genes manually. Important: Additionally, if you have used the "Input Quick Gene Selection List" to quickly select a set of genes, further changes made by manually selecting/deselecting genes will mean that the gene selection no longer follows what was specified in "Input Quick Gene Selection List". If you want to reset the gene selection to what was provided in the textbox, simply click the "Select Genes From Text Input" button again. When you are done, click "Save and Continue". back to top 1.4: Motif SettingsThere are 3 sections on this page. The first section controls "Exclusion Sequences". In some circumstances, you may want the output sequence to exclude certain specific nucleotide sequences. E.g. Perhaps you are going to splice this sequence into a vector using a certain restriction enzyme, and hence you do not want the enzyme's target site to occur within the output sequence. We provide this optional facility, where you may enter nucleotide sequences which you want to be excluded from the output, as a comma seperated list. This page also has a dropdown menu bar, which allows you to select common Restriction Enzyme and Translation Initiation Sites by name. Clicking on a site name, will add its sequence to the textbox. For Restriction Enzyme Sites we also include the complement (since Restriction Enzymes work at the DNA level which ignores directionality). But for the Translation Initiation Sites, we do not include the complement (since Translation Initiation works at the mRNA level, and it usually does not matter if the mRNA complement has a Translation Initiation Site). For the Translation Initiation Sites, we used the following consensus sequence (a particular base was excluded from a specified position, if it occurred less than 10% of the time):
This dropdown menu requires Javascript to work, so if Javascript is not fully enabled, it may fail to function. The second section determines whether you want to enable removal of "Consecutive Repeats". You can specify both the "Length of Nucleotide Motif", and the "Minimum Number of Instances before removal". The "Length of Nucleotide Motif" refers to the number of nucleotides within one iteration of the repeat. E.g. If your "Length of Nucleotide Motif" is 2, this will include all possible 2^4 = 16 dinucleotide motifs (AA, AT, AC, AG, TA, TT, TC, TG, CA, CT, CC, CG, GA, GT, GC, GG). The "Minimum Number of Instances before removal" indicates how many times the motif should occur, before it is considered a repeat that should be removed. This parameter has a minimum value of 2. E.g. If "Minimum Number of Instances before removal" is 3, then a particular motif must occur 3 or more consecutive times to be considered a repeat that should be removed. Hence "ATATAT" would qualify, but "ATAT" would not. The third section determines whether you want to enable removal of "Repeated Motifs". Whereas the "Consecutive Repeats" specified in the previous section have to be next to each other, "Repeated Motifs" can have each instance of the repeat located anyway in the sequence. "Length of Nucleotide Motif" has a minimum of 7, and "Minimum Number of Instances before removal" has a minimum of 2. E.g. "ATGATGATGCTCATGATGATGCGCATGATGATG" would count as a 9 base long motif, that has 3 instances. Important: If you do want to enable removal of "Consecutive Repeats" or "Repeated "Motifs", ensure that their respective checkbox is ticked. Otherwise the algorithm will ignore whatever values you may have entered. When you are done, click "Save and Continue". Note that the algorithm is NOT guaranteed to remove all exclusion sequences and/or repeats. It will try its best to exclude them, but there may be instances where this is simply not possible for the given protein sequence. E.g. Lets say that the protein has 2 consecutive methionine bases. This can only be coded in one way (ATGATG) under standard translation rules. If you specify "ATGATG" as an exclusion sequence, or that you want to remove repeats of 3 nucleotide motifs repeated 2 or more times (which includes ATGATG), the algorithm will be unable to meet your request. As such, the output results include count checks on whether any Exclusion Sequence or User-specified Repeats are present in the output sequence. Use these to double check whether all Exclusion Sequences and Repeats are absent. back to top 1.5: Submit JobThis is the last page for setting up a job. Optionally, you may enter your e-mail address, and this web tool will send you a notification e-mail, when your results are ready. Regardless of whether you enter an e-mail, when you are ready to continue, click "Save and Submit Job". Important: Once a job has been submitted, you cannot make any other further changes to the input parameters. This page is hence the final confirmation before the algorithm starts running. back to top 2: Waiting for ResultsImmediately after you submit a job, you are brought to the results page. If your job is not yet finished (as will likely be the case when you first come here), it will display a "Please Wait" message. Your results are tied to the URL address of this page. Hence, you may bookmark or otherwise note down the page address, to check on your results at a later time. The page has a hidden timer that refreshes the page every minute. So you may leave the page open, and it will refresh itself, until your job is finished, at which point it will display the results. Note however, that this refresh timer requires Javascript to work, so if Javascript is not fully enabled, it may fail to function. back to top 3: Sample DataWe have provided an option for users to generate a sample output dataset, so that they may view an example pareto spatial distribution, without having to go through the whole job submission process. This sample data was generated using the following inputs and parameters:
For help on how to intepret this sample data, see the next section below. back to top 4: Intepreting ResultsIn the following sections, we explore how to understand the results that this website gives you. But firstly, a quick note on how the results are generated. This website uses a genetic algorithm, the details of which can be found in our previous publication. What follows is a simplified summary:
If the job has only one optimization parameter, then the fittest sequence will be the one with the highest fitness value for that one optimization parameter, and hence there will only be one output sequence. But if there are multiple optimization parameters, multiple output sequences can be considered to have approximately the same overall fitness value, but with different values for the different fitness parameters. In this case, the results will contain ALL of these output sequences, and we let the users select one which best fits their requirements. E.g. Lets say that we have 2 optimization parameters. We are trying to maximize both Individual Codon Usage and Codon Context. The algorithm will produce a range of output sequences. At one extreme, there will be a sequence with very high Codon Context (CC) fitness, but very low Individual Codon (IC) Fitness (top left point in the graph below). There will be other sequences with increasing IC fitness, but decreasing CC fitness (curve decreases as it moves rightwards). Until we reach the other extreme, with a sequence which has very high IC fitness, but very low CC fitness (bottom right point). 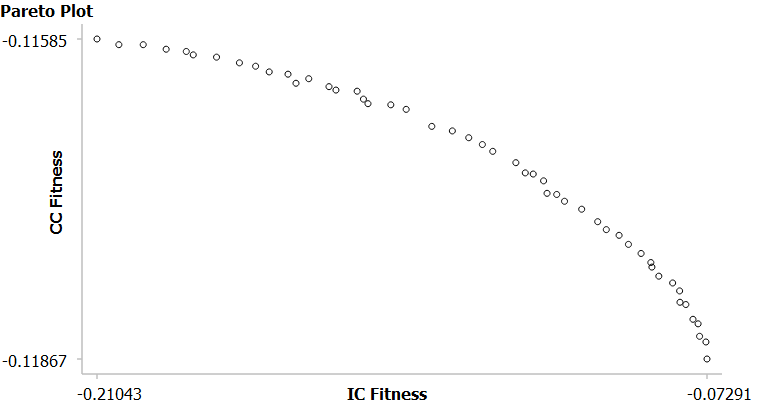
Note that the difference between "Maximize" and "Minimize" for a parameter, is based on how fitness is calculated, but only during the evolution process. When "Minimize" is selected, the algorithm simply calculates the "Maximize" fitness value, and then takes the negative. In this way, sequences which have the highest value under "Maximize", will have the lowest value under "Minimize". However, this only applies within the evolution process- the fitness value provided in the final output reports will be based on the "Maximize" calculation, regardless of whether "Maximize" or "Minimize" was selected. back to top 4.1: Summary Graph and TableWhen the algorithm has finished processing your data, it will first show you a summary of all the output sequences of the final generation. (See the section above to understand why there may be one or multiple output sequences). But before the summary, there will be 2 links at the top of the page. The first lets you add or remove User Defined Sequences (see this section for more details). The second lets you export all the results to a tab seperated TXT file, which shows all the output sequences, and their associated fitness values. Important: Jobs will be deleted approximately one month after they were submitted. If you wish to preserve your results, please save it as a TXT file or PDF document. If you have at least 2 of the following Optimization Criteria (IC fitness, CC fitness, CAI, Hidden Stop Codons, 5' RNA folding instability, Codon Auto-Correlation, GC Content Fitness) the page will also display a graph. This is called a Pareto Plot, but due to the constraints of web interfaces, is limited to only 2 dimensions (instead of however many Optimization Criteria you may have specified). If 3 or more Optimization Criteria were specified, since the graph is compressed onto 2 dimensions, the points may not exhibit a smooth curve, that is typical of Pareto distribution, as the points follow a hyperplane being projected onto 2 dimensional space. Besides the basic 2 dimensions of X and Y axis, the graph can also a indicate a 3rd dimension by colour coding the points. This website uses cyan-red colour coding, which should be readable by most colour blind users. Some of the fitness values appear as negative numbers. In such cases, the closer the negative value is to zero, the higher the fitness of the sequence. For IC fitness, CC fitness, Codon Auto-Correlation and GC Content Fitness, the values refer to how far the output sequence differs from your specified target. (E.g. assuming a target GC content of 50%, an output sequence with a GC content of 40%, and another with a GC content of 60%, will both have the same GC Content Fitness of -0.1, indicating a 10% deviation from the desired GC content). In the case of 5' RNA folding instability, the value indicates the mRNA mean folding value (MFE) values, as computed using Vienna RNA. Hence the closer this value is to zero, the greater the predicted instability of the 5' RNA, and the higher its fitness. Each datapoint on the graph represents an output sequence. Mousing over a datapoint will bring up its fitness value details. These values will appear in one or both of the following:
Exactly where the values appear depends on Browser compatability. The page has been designed and tested so that at least one of the above will work on most major browsers. Clicking on the output sequence brings you to its details page (see the section on result details for more information). Note that there are 2 types of datapoints: The optimized sequences generated by our algorithm, and custom sequences which can be added or removed by the user (see this section for more details). The user defined sequences use a different symbol (triangle instead of circle) to help them stand out. Immediately below the graph are a set of dropdown boxes. Here, you may select which Optimization Criteria, you want to be plotted on which axis, as well as what values you want to use to colour code the points. Once you have selected your desired options, click the "Redraw Graph" button to the immediate bottom right of the dropdown boxes, to generate a new graph. Note: Because colour coding is not a true axis dimension, we also allow you to plot values, which are not among the normal pareto fitness parameters meant for the X/Y axis. Hence, beyond the usual Optimization Criteria, you may also choose to plot: the GC content (not just the fitness), the number of Exclusion bases, the number of Consecutive Repeat bases, or the number of Repeated Motif bases. (E.g. This can be useful if you want to visualize which of the output sequences have fewer number of repeated motifs). If you have only 1 Optimization Criteria, then the graph (and the dropdown boxes) will be absent. You will only have the summary table instead. Since the algorithm only reports the fittest sequences, if there is only 1 Optimization Criteria, the fittest sequence will be easy to determine, and there will usually only be 1 output sequence as well. The summary table lists the fitness values for each output sequence (both the optimized sequences generated by our algorithm, and the user defined sequences). It is similiar to the exported TXT above, except that it does not include the actual output sequence due to space constraints. This table can be sorted by clicking on its headers, and you can sort by multiple columns, by holding shift while clicking on each header. Clicking on the name of a sequence will lead you to the details page for that output sequence (see the section below). back to top 4.2: Add/Remove User Defined SequenceSometimes you might want to compare the optimized nucleotide sequences generated by our algorithm, with optimized sequences obtained from other sources. To facilitate this, we allow users to add their own custom User Defined Sequences (UDS) to the list of output sequences. We will calculate the corresponding fitness values for your UDS, and add them to the summary table (and display them on the summary graph as well, if there are at least 2 Optimization Criteria). The "Add/Remove User Defined Sequence Page" is the central point from which you may manage your list of UDS. At the top of the page is the "Add New User Defined Nucleotide Sequence to Results" section. You can add a UDS here, by simply pasting your nucleotide sequence into the textbox, and clicking the "Add Sequence" button (or the "Add And Return to Summary" button). Optionally, you may also specify a name for the UDS, so that you can tell multiple different UDS apart. You can add up to 10 UDS to each job, and once you reach that limit, this section becomes disabled. Below that is the "Delete Existing User Defined Sequences" section. Here, the website displays a table with your current list of UDS, along with their fitness values. You can delete a UDS, simply by clicking on the "Delete" link on the far right column. back to top 4.3: Detailed ResultsWhereas the results summary page gives a quick overview of all the output sequences (as described in the section above), this page provides in depth details for one particular sequence. The exact items that are included in your results will depend on the options you selected for a particular job. Just below the page title, there will be a link that leads you back to the results summary page. Below that will be a Summary of the fitness values for this particular output sequence. Next to the Summary title, is a link to the results as a downloadable PDF document. The PDF will have all the same information as this results page. But since PDF is a static document, it will lack some of the dynamic display options that the webpage has (e.g. show/hide functions and sortable tables). Important: Jobs will be deleted approximately one month after they were submitted. If you wish to preserve your results, please save it as a TXT file (above section) or PDF document. Below the summary is a textbox with your output sequence translated into a protein. You may use this to double check that your output sequence encodes the correct amino acids. Below the protein, is another textbox with your actual nucleotide output sequence. Clicking on either of these textboxes will automatically select the entire sequence, and from there you may copy and paste it to other places. Next there may be the section "Output Sequence Relative Frequency Distribution". This section appears if you have selected at least one of the following Optimization Criteria: Individual Codon Usage, Codon Context or Codon Adaptation Index. This shows the output sequence, but with colour coding indicating the relative frequency. "Relative frequency" refers to the frequency of a codon (or codon pair) in comparison to other synonymous codons (or codon pairs). It is derived by dividing the raw count of that codon, by the synonymous total of all codons which encode the same amino acid. (Both raw count and synonymous total refer to the count within the target expression host, rather than for the output sequence alone.)
The exact mode of display will depend on the Optimization Criteria you have selected:
For Codon Adaptation Index, the objective is to always use the highest frequency synonymous codons. But for Individual Codon Usage and Codon Context, the goal of the algorithm is NOT to simply use the highest frequency synonymous codon (or codon pair) at all times. Instead, it tries to mimic the target species natural codon frequency. So there will still be low frequency codon (or codon pairs) appearing on occasion. Below the "Output Sequence Frequency Distribution", there may be an "Exclusion Sequence Fitness" section. This will only appear if you provided a list of exclusion sequences. As a quick summary, the section title notes the total number of nucleotide bases that fall under an Exclusion Sequence. In most cases, there should be no exclusion sequences (i.e. Total Exclusion Bases: 0), and you should not need to investigate further. However as noted above, the algorithm is not guaranteed to remove all exclusion sequences, so always check this to make sure. This section is further sub-divided into 2 reports. The first report is a table which lists each individual exclusion sequence, and the number of times it has been found in the output sequence. The second report is the output sequence, but with exclusion sequences highlighted and in bolded capital letters. Next is the "Consecutive Repeat Fitness" section. This will only appear if you enabled "Consecutive Repeat" removal. As a quick summary, the section title notes the total number of nucleotide bases that are part of a "Consecutive Repeat" sequence (if it is '0', then you do not need to check further). The report consists of the output sequence, but with "Consecutive Repeats" highlighted and in bolded capital letters. Because of the nature of repeated sequences, there may be some instance of overlaps, where 2 or more sets of "Consecutive Repeats" share some of the same nucleotide bases. E.g. Lets say we enable repeat removal of "Consecutive Repeats" with a motif length of 3, repeated 3 or more times. Consider the sequence "ATCGTCGTCGTCA". This actually has multiple "Consecutive Repeats" overlapping each other: From "ATCGTCGTCGTCA" to "ATCGTCGTCGTCA". Overall, "ATCGTCGTCGTCA" will be flagged for removal. After that is the "Repeated Motif Fitness" section. This will only appear if you enabled "Repeated Motif" removal. As before, the section title notes the total number of nucleotide bases that are part of a "Repeated Motif" (if it is '0', then you do not need to check further). The report consists of the output sequence, but with "Repeated Motifs" highlighted and in bolded capital letters. As with "Consecutive Repeats", there may be some instance of overlaps, where 2 or more sets of "Repeated Motifs" share some of the same nucleotide bases. E.g. Lets say we enable repeat removal of 7 nucleotide motifs, repeated 2 or more times. Consider the sequence "ATGATGAGGCCCATGATGAGG". This actually has multiple "Repeated Motifs" overlapping each other: From "ATGATGAGGCCCATGATGAGG" to "ATGATGAGGCCCATGATGAGG". Overall, "ATGATGAGGCCCATGATGAGG" will be flagged for removal. If Individual Codon Usage Or Codon Adaption Index was one of the optimization criteria, next up there will be a bar chart titled "Host Vs Optimized Codon Relative Frequency Chart". This compares the usage pattern for each codon, between the host (red), and this current output sequence (blue). If the input sequence was DNA/RNA, then the next section will be a base-by-base comparison of the input nucleotide sequence, against the current output. The comparison will mark any changed bases, as either Transversions (purine to pyrimidine or vice versa) or Transitions (purine to purine or pyrimidine to pyrimidine). Finally we have the Frequency and Colour tables. This section appears if you have selected at least one of the following Optimization Criteria: Individual Codon Usage, Codon Context or Codon Adaptation Index. There maybe one or two tables, depending on the Optimization Criteria you have selected:
Each table shows a list of codons (or codon pairs), and its corresponding Host Relative Frequency (i.e. the relative rate of occurence for that codon, compared to other synonymous codons which code for the same amino acid, within the expression host). The cell background colour will be the same colour used in the "Output Sequence Relative Frequency Distribution" above. It also shows the Optimized Relative Frequency, and detailed calculations on how this value is derived (Synonymous Total is the sum of Raw Count for all the synonymous codons. Relative Frequecy is the Raw Count, divided by the Synynomous Total). These tables are sorted by amino acid by default, to group synonymous codons (and/or codon pairs) together. But the tables also come with sortable headers, so you may sort by alternate criteria if you wish. Additionally, you may sort by multiple columns at once, by holding down the 'shift' key while clicking on each header. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
